Don’t you know how to use dnastar to quickly calculate the GC content of DNA sequences? If you don’t know how, come and learn with the editor how to use dnastar to quickly calculate the GC content of DNA sequences. I hope that through this tutorial, you can help everyone better understand the dnastar software.
Step 1: Download and install dnastar on your computer. After the installation is complete, select the EditSeq program. The functions of this program are to analyze GC content, find bases at a certain position, base proofreading, translate DNA sequences into amino acid sequences, sequence reverse complementation, sequence reverse, etc.

Step 2: Copy and paste the DNA sequences you want to analyze, then select all DNA sequences. Note that if unchecked, the corresponding option cannot be applied.
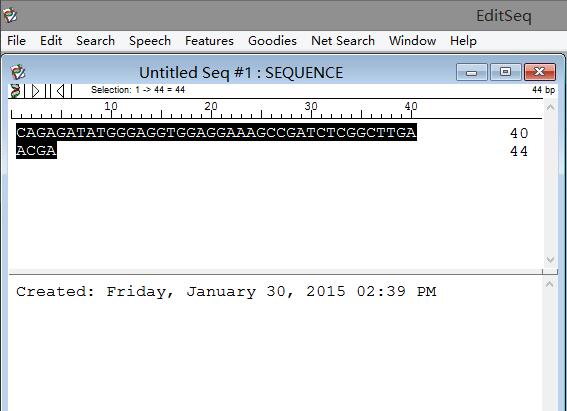
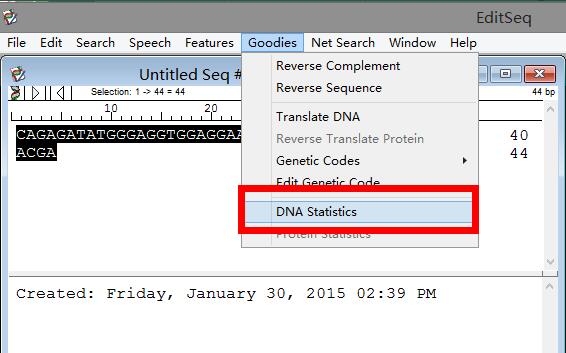
Step 3: Click "Goodies" > DNA statistics in the title bar, as shown in the figure below:
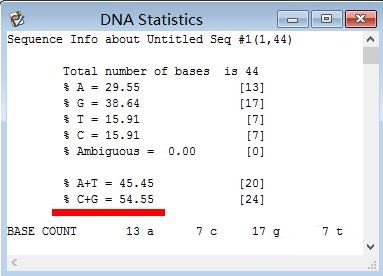
Step 4: Then you can see the GC content in the pop-up window, and even the ATGC content of each base can be accurately calculated.
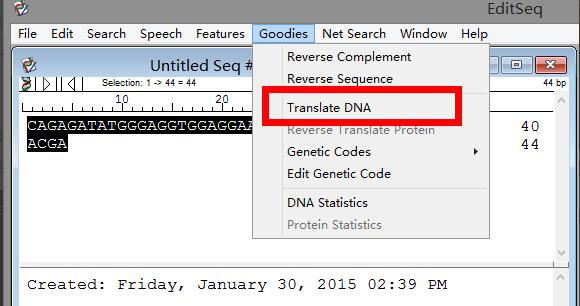
Return to step 3. You can see "Translate DNA" on DNA statistics. After clicking it, you can see the translated amino acid sequence in the pop-up window.
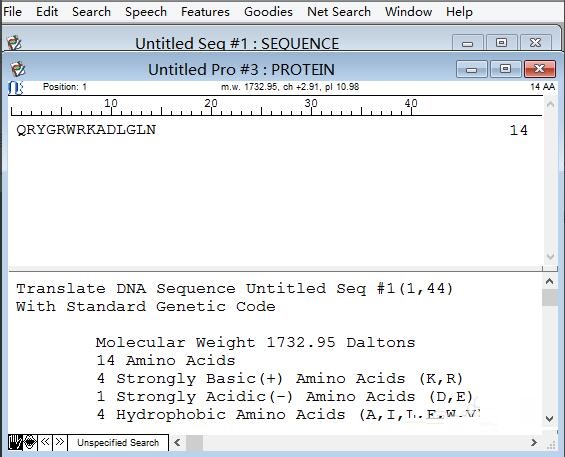
The above is the dnastar method shared by the editor to quickly calculate the GC content of DNA sequences. Friends in need can take a look.




